About the Department of Systems Biology
One of the defining features of living organisms is their astonishing complexity. Even seemingly simple single cell organisms such as microbes display exceedingly complex behaviors, determined by intricate molecular networks in which large numbers of molecular components, pathways and chemical reactions act together. These behaviors have fascinated scientists for decades and include development, response to pathogenic and environmental insults and interactions with other organisms. Understanding how complexity of living systems arises and coordinates cellular function and pathologies continues to be one of the principal goals of biomedical research today. Read more about how the Department of Systems Biology tackles these questions on our Research and About pages.
The Department of Systems Biology (DSB) studies how biological complexity can be derived and understood from the interplay between individual components and processes that make up living organisms.
For information about our Graduate and Summer Undergraduate Programs as well as the application process, please see our Education Page.
DSB Spotlight

A new paper published in Nature Cell Biology from the Dekker lab identifies two distinct folding programs – one dependent on chromosome intrinsic factors and the other on imported cytoplasmic factors – necessary for establishing interphase chromosome conformation as cells proceed from mitosis to G1. Learn more about this work, highlights from the experimental journey and the first author Allana Schooley in this Q&A.
Read the paper: Interphase chromosome conformation is specified by distinct folding programmes inherited through mitotic chromosomes or the cytoplasm
Read Accompanying News & Views: Inheriting chromosome conformation
DSB Seminar Series
|
All seminars will take place at 11am in AS6-2072 (unless otherwise noted) |
| John Doench, PhD |
| Director of Research and Development in the Genetic Perturbation Platform, Broad Institute |
| Title: "Where Does He Get Those Wonderful Toys? A Tour of the Functional Genomics Toolbox" |
| January 29, 2026 |
| Host: Jolanda van Leeuwen |
| Stirling Churchman, PhD |
| Professor, Harvard Medical School |
| February 12, 2026 |
| Host: Marian Walhout |
| Alison Taylor, PhD |
| Assistant Professor, Columbia University Medical Center |
| February 19, 2026 |
| Host: Emma Watson |
Recent Publications
|
Bacterial population dynamics during colonization of solid tumors |
| Molecular Systems Biology. 2025 Dec 15 |
| Serkan Sayin, Motasem ElGamel, Brittany Rosener, Michael Brehm, Andrew Mugler, Amir Mitchell |
|
|
| Nature Cell Biology. 2025 Dec 22 |
| Allana Schooley, Sergey V Venev, Vasilisa Aksenova, Jesse W Lehman, Emily Navarrete, Athma A Pai, Job Dekker |
| Also see accompanying News & Views: Inheriting chromosome conformation |
|
An integrated view of the structure and function of the human 4D nucleome |
| Nature. 2025 Dec 17 |
| Job Dekker, Betul Akgol Oksuk,...., Liyan Yang, Johan H Gibcus,....Sergey V Venev |
| Also see accompanying News & Views: Systematic maps reveal how human chromosomes are organized & Research Highlight from UMASS Chan News: 4D Nucleome Consortium produces detailed models of the 3D genome over time in cells |
|
E. coli transcription factors regulate promoter activity by a universal, homeostatic mechanism |
| Science. 2025 Sept 11 |
| Vinuselvi Parisutham, Sunil Guharajan, Melina Lian, MD Zulfikar Ali, Hannah Rogers, Shannon Joyce, Mariana Noto Guillen, Robert C Brewster |
|
Predicting drug inactivation by changes in bacterial growth dynamics |
| npj Antimicrobials and Resistance. 2025 Sept 9 |
| Carmen Li, Serkan Sayin, Ethan Hau Chian Chang, Amir Mitchell |
|
RNA Pol II inhibition activates cell death independently from the loss of transcription |
| Cell. 2025 Aug 15 |
| Nicholas W. Harper, Gavin A. Birdsall, Megan E. Honeywell, Kelly M. Ward, Athma A. Pai, Michael J. Lee |
|
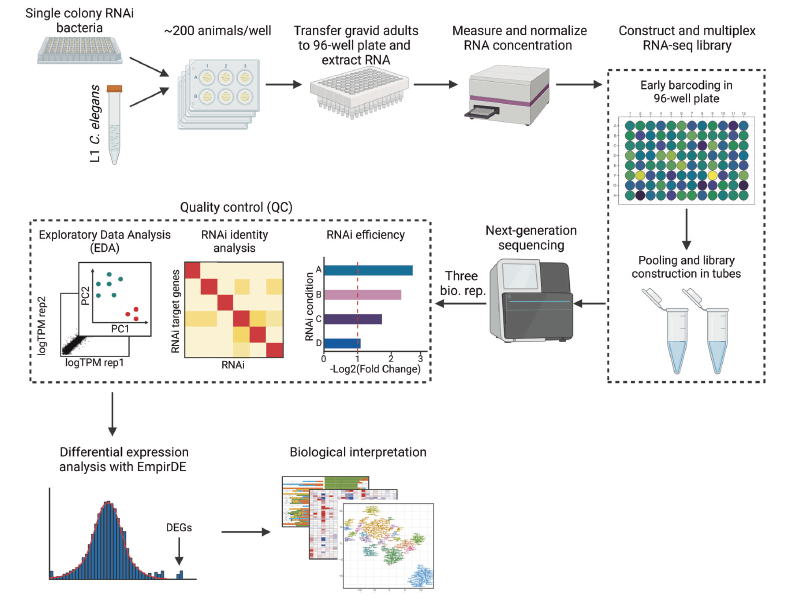
Worm Perturb-Seq: massively parallel whole-animal RNAi and RNA-seq |
| Nature Communications. 2025 May 23 |
| Hefei Zhang, Xuhang Li, Dongyuan Song, Onur Yukselen, Shivani Nanda, Alper Kucukaral, Jingyi Jessica Li, Manuel Garber, Albertha J.M. Walhout |
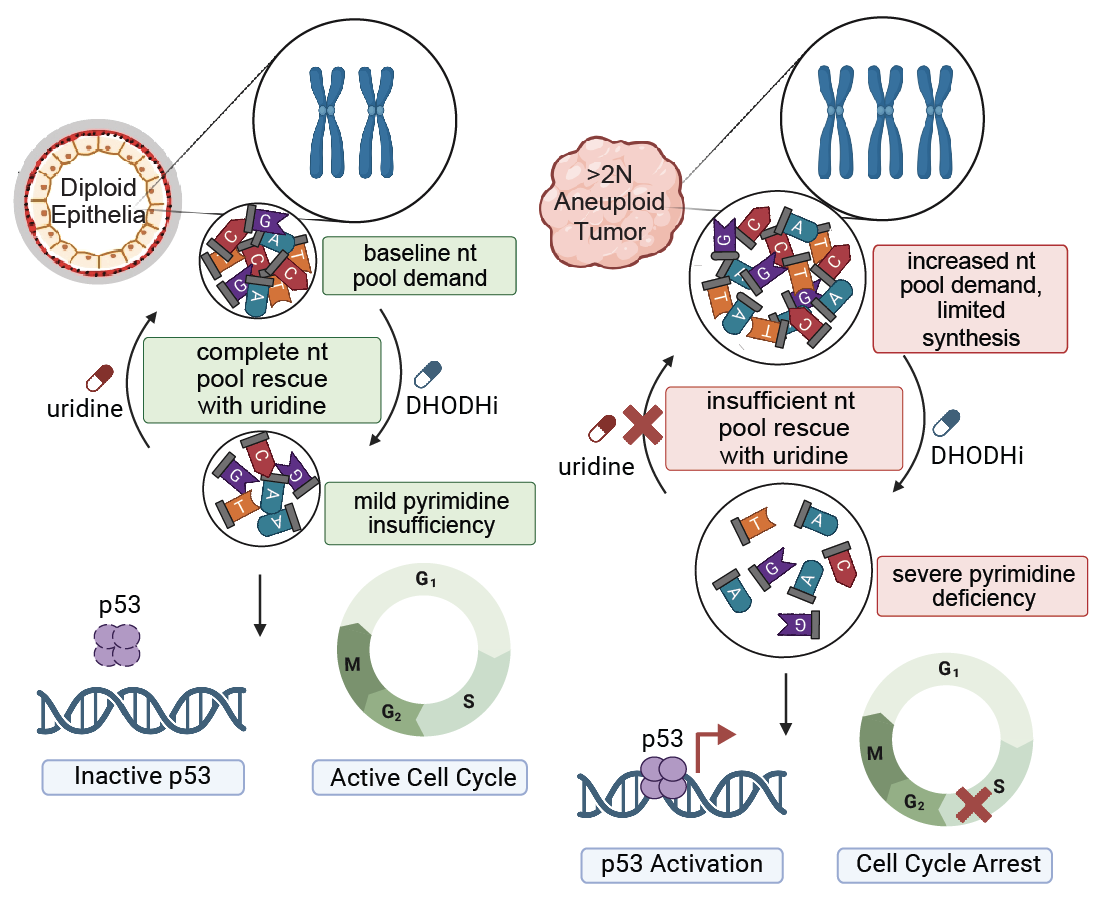
Aneuploidy generates enhanced nucleotide dependency and sensitivity to metabolic perturbation |
||
| Genes & Development. 2025 May 5 | ||
| Rayna Y. Magesh, Arshia N. Kaur, Faith N. Keller, Abdulrazak Frederick, Tenzin Tseyang, John A. Haley, Alejandra M. Rivera, Anthony C. Liang, David A. Guertin, Jessica B. Spinelli, Stephen J. Elledge, Emma V. Watson |